Genetically-encoded Sparse Cell Labeling – A SPARC of Innovation
Until recently, there were no completely genetic tools that would allow researchers to label just a fraction of a single genetically-defined subset of cells. By labeling fewer cells in a population, it’s easier to visualize individual/non-overlapping cells. While transgenic animals are commonly used to specifically manipulate a cell type of interest in an organism, all cells of that type are affected. Previous tools to overcome this restraint include an AAV-based sparse labeling system where a limiting amount of AAV are injected into the brain of Cre-expressing mice to trigger recombination and expression of a transgene (Lin et al., 2018). However, this system requires precise titration of the AAV. So far, tools to overcome this challenge in Drosophila require heat-shocking the system or using chemical inducers of gene expression (del Valle Rodríguez et al., 2011).
Genetics of SPARC
| Figure 1: SPARC and SPARC2 use the GAL4-UAS system along with precisely truncated attP sites to control recombination of the gene construct. A predictable number of cells will express the effector or retain the stop cassette following recombination. Image from Isaacman-Beck et al., 2019. |
Sparse Predictive Activity through Recombinase Competition (SPARC), a genetic tool that allows for the labeling of a subset of cells with the same genetic makeup, overcomes all of these labor-intensive and invasive techniques to reliably label any post-mitotic cells in Drosophila. SPARC was developed by Thomas Clandinin’s lab, and incorporates several common genetic tools in Drosophila (Fig. 1):
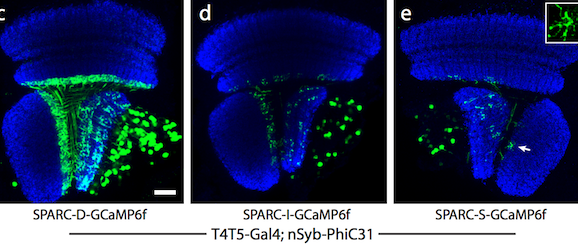
To more precisely control the amount of labeling, you can alter the attP site. Truncating the attP site upstream of the stop cassette results in decreased efficiency of recombination and preferentially leads to gene constructs where the stop cassette remains following recombination. This strategy results in sparse labeling of the cell population that expresses SPARC and the fraction of cells that will be labeled in a given population is directly correlated to the length of the truncated attP site. The three SPARC variants, based on the length of the attP site, allow for dense (D), intermediate (I), or sparse (S) labeling of the cell population.
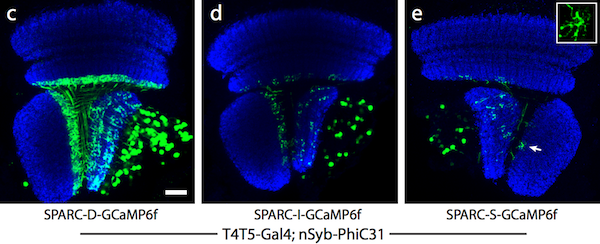 |
| Figure 2: Dense (c), intermediate (d), and sparse (e) labeling of the calcium indicator, GCaMP6f, in the Drosophila optic lobe. Image from Isaacman-Beck et al., 2019. |
Applications for SPARC
SPARC technology is versatile and can be applied across cell types and organisms by replacing the transcription factor binding site and effector in the construct. In Drosophila, the SPARC systems can be used both for visualization and for optogenetics. The Clandinin lab used SPARC to image the calcium response in the dendrites of T5 neurons by placing the calcium indicator GCaMP6f in the effector position, as well as to optogenetically control the depolarization of R2 neurons using CsChrimson::tdTomato, an optogenetic tool (CsChrimson) attached to a fluorescent indicator (tdTomato), as the effector. They also expanded the SPARC system to use other experimental transcription activation systems, such as LexA/p65 or mCD8/GFP, to allow sparse labeling in a variety of fly cell types. Because the PhiC31 recombinase can work in other model organisms including mouse and fish, SPARC is likely generalizable to other systems as well.
References:
Additional resources on the Addgene blog
