An inexpensive paper-based test that can rapidly identify multiple types of dangerous bacteria – Innovation Toronto
Bacterial infections are the leading cause of disease and death worldwide; an ongoing public health problem exacerbated by slow or inaccurate diagnostics. Now NIBIB-funded scientists have engineered an inexpensive, paper-based test that can rapidly identify multiple types of bacteria.
The research team at the University of Nebraska used a complex blend of microbiology, chemistry, and artificial intelligence (AI) to create a testing platform that appears deceptively low-tech—built for use in remote low-resource environments such as field hospitals and rural clinics.
“We have designed this technology to be extremely sensitive and accurate for identifying bacterial species while also being easy to manufacture,” explained Denis Svechkarev, Ph.D., research assistant professor in the department of pharmaceutical sciences and co-first author of the paper with graduate student Aayushi Laliwala. “The test is also durable enough to survive shipping to remote locations and simple enough to be easily used by healthcare personnel with limited training and equipment.”
The work is being conducted in the laboratory of Aaron M. Mohs, Ph.D., associate professor in the department of pharmaceutical sciences and senior author on the publication, which appeared in the journal Analytical Chemistry1 on January 24.
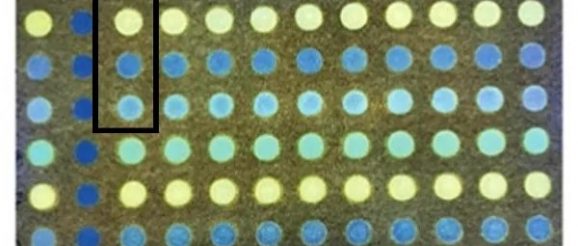
The “simple” platform, which is in the development and testing stages with the hope of eventual use in the field, has a complex name, “Paper-Based Ratiometric Fluorescent Sensor Array.” About the size of a 3 x 5 card, the paper sensor is “arrayed” with a grid of small circles on which the bacterial specimens to be tested are applied. The “ratiometric fluorescent” part of the name refers to the ingenious way the bacteria are identified.
The research team designed and synthesized fluorescent dyes that can “sense” the subtle biochemical differences of each type of bacterium and turn those differences into different fluorescent signals. Four different fluorescent dyes are dried onto four circles on the array comprising a single test. A bacterial specimen, such as e coli is placed on each of the four circles and the dyes are activated with ultraviolet light resulting in the four dyes each sending out five fluorescent signals for a total of 20 fluorescent signals per test.
A fluorescent plate reader scans the 20 fluorescent signals, which vary depending on the interaction of the dyes with the outer membrane of the bacteria. A state-of-the-art AI program—in the form of an artificial neural network—was trained to recognize the subtle but specific pattern of fluorescent intensities created by each type of bacteria. The result is a “signature” fluorescent pattern that is transferred from the reader into the artificial neural network program, which identifies the type of bacteria.
In collaboration with the microbiologists, Drs. Marat R. Sadykov and Kenneth W. Bayles, the team tested the system using a collection of 16 bacterial species. The system correctly identified the 16 species more than 90% of the time—a level of accuracy that could provide a healthcare worker in the field valuable information about the specific bacteria in an infected individual allowing for precise, prompt antibiotic treatment. The test also determined whether the bacteria was gram positive or negative with 95% accuracy. Gram testing is a technique that further determines the makeup of bacteria and is critical to knowing which types of antibiotics are most effective. The accuracy of the test was extremely promising considering that a few hours of delay in diagnosing and treating an infectious disease dramatically worsens the patient’s prognosis.
Every aspect of the test was designed for potential use in even the most remote parts of the world, where current techniques that require sophisticated equipment and expertise are not feasible. For example, drying the fluorescent dyes onto the paper card removed the need to use liquid fluorescent dyes that would require refrigeration—often unavailable in low-resource regions. Photolithography was used to “photo-stamp” the grid of circles onto the paper card—a rapid and inexpensive way to manufacture thousands of cards. In testing—done by placing the cards in a box in the closet—the cards remained stable for up to 6 months, making them ideal for shipping and distribution to remote areas. The pattern on the card is identical to the 96 well plates used for many tests that use liquid components, which allows the paper cards to be scanned and read by readily available standard machines.
“This project is an extraordinary example of how making something simple requires the use of multiple complex technologies,” said Tatjana Atanasijevic, Ph.D., (Scientific Program Manager) of the program in Bioanalytical Sensors at the National Institute for Biomedical Imaging and Bioengineering (NIBIB), which co-funded the project along with several additional institutes at the National Institutes of Health.
The work is in the research and development stage and the team is testing and refining the system using samples that replicate what would be collected from patients in the field. Future technical feats in the team’s crosshairs include working with engineers to create a system that would allow the paper 96-spot card to be read with a simpler device, maybe even a cell phone camera—an admittedly lofty goal, but doable explained Svechkarev.
Asked about the work, Mohs credits the extraordinary effort of Svechkarev and Laliwala. “The technology needed to create this bacterial detection system was devised during the pandemic when we had limited access to the laboratory. Denis and Aayushi used this time to develop skills that included new computer coding methods, learning how to use different types of artificial intelligence, and finalizing the design of the best fluorescent dyes—all key elements that came together to build this promising diagnostic system.”
More from: University of Nebraska
