How machine learning is driving innovation in cardiovascular disease research
One day in 2019, cardiologist Patrick Ellinor was leaving his office at the Broad Institute of MIT and Harvard, deep in thought. He’d just become head of the Broad’s Cardiovascular Disease Initiative and knew it was time to sketch out a plan for the future of cardiovascular research at the Broad.
As he was walking down the hall, he ran into Anthony Philippakis, a geneticist and statistician trained in cardiology who is now Broad’s chief data officer. The two started talking about the Data Sciences Platform at the Broad, which Philippakis led at the time — how it was full of driven researchers developing software, but didn’t have a strong clinical motivation. They contrasted that team with scientists in cardiovascular research, who had experience studying disease, but lacked the computational skills to deeply analyze biomedical data and gain new insights into how to better diagnose and treat heart disease.
 Patrick Ellinor
Patrick EllinorCredit: Maria Nemchuk
An idea soon emerged: bridge the two fields and kickstart collaboration by encouraging each group of scientists to learn the other’s language. Philippakis, Ellinor, and their colleagues decided, as a first step, to host a hackathon. They invited data scientists and cardiovascular disease researchers, gave them genetic and medical data from the UK Biobank, and asked them to answer — together — pressing clinical questions such as how to predict which patients would develop atrial fibrillation, a condition involving irregular heart rhythms.
“It was pretty amazing,” remembers Ellinor, who is also an institute member at Broad, acting chief of cardiology at Massachusetts General Hospital (MGH), and a professor of medicine at Harvard Medical School (HMS). “In just a couple of weeks, the different teams were able to parse the data, talk together, and even recapitulate a lot of what we know about cardiovascular disease, epidemiology, and disease drivers.”
The hackathon also showed that the two groups could and were excited to work together.
 Anthony Philippakis
Anthony PhilippakisCredit: Maria Nemchuk
“There was a real sense of optimism,” Philippakis said. “We saw that this was an amazing opportunity, both as a way of approaching tough challenges in medicine and a way of bringing together a great group of people.”
Soon after the hackathon, Ellinor, Philippakis, and other scientists from the Cardiovascular Disease Initiative founded the Machine Learning for Health (ML4H) team, which contains Broad researchers as well as scientists from MGH, Brigham and Women’s Hospital, and MIT. ML4H applies machine learning methods — which detect patterns in data to make predictions about new data — to accelerate the study of disease, improve clinical trials, and support clinical decision making.
Nearly three years later, ML4H has grown to include nine principal investigators and more than 30 other researchers: computer scientists, biologists, and physicians. Ellinor and Philippakis are more excited than ever about the community they’re building. Their collaborative teams have shown, in a flurry of studies in recent years, how machine learning and other computational tools have helped them find new genes and markers for a variety of cardiovascular diseases, which could one day enable doctors to run more efficient clinical trials, better identify high-risk patients, and select the right treatments.
“Machine learning is perfectly suited to identify subtle features of disease in high resolution data (like imaging) beyond what clinicians can see with their own eyes,” said Puneet Batra, who was director of machine learning in the Data Sciences Platform and leader of the ML4H team until 2022. “Machine learning gives us a new window into the richness of data,” he said. “And that is bringing us closer to the dream of precision medicine — predicting which individuals will get sick, and how we can prevent that from occurring or heal them.”
A widespread need
Cardiovascular disease is the leading cause of death worldwide. In the US, medical interventions such as surgery, implants, and cholesterol-lowering drugs have helped drive down death rates, but that decline has slowed in recent years, highlighting a need for new research approaches.
Cardiovascular diseases, however, can be challenging to study. The complex mix of genetic and environmental factors such as a diet and exercise are difficult to untangle and study in animal models. Clinical trials testing new heart drugs are expensive because they must monitor large numbers of people until the disease, such as stroke, develops. New treatments must also have few side effects since patients typically take them for the rest of their lives. As a result, Ellinor says, it can be difficult to translate biological insights into new therapies.
At the same time, cardiologists have plenty of tools at their disposal to measure and study heart structure and function.
new window into the richness of data.
“Cardiology is a great field to apply the tools of machine learning to, because the heart is one of the organs that we can best phenotype,” said Philippakis. He notes that doctors have a suite of imaging tools at their disposal, ranging from magnetic resonance imaging (MRI) and electrocardiograms to angiograms and positron emission tomography scans. These techniques can help doctors diagnose and monitor heart problems such as torn valves, blocked arteries, and irregular heart rhythms in patients. But all this clinical data, when pooled together from large numbers of patients, can also be mined by researchers to shed light on the biological underpinnings of these diseases.
“Each of these modalities gives us very rich, high dimensional data sets — which are exactly what we need to build machine learning models that both predict who will develop disease and who will most respond to therapies,” Philippakis said.
Hidden patterns
One particularly informative type of data from heart patients is MRI, which uses magnetic fields and radio waves to give doctors a real-time picture of a patient’s beating heart, revealing a wide range of features such as thickened muscles and impaired blood flow. Broad researchers have set out to develop machine learning models that can automatically analyze large numbers of MRI images and extract key features that are linked to various diseases, in hopes of then finding new genetic factors associated with those diseases.
In 2021, James Pirruccello, then a junior faculty member at HMS and MGH and a researcher on Ellinor’s team, applied this approach to a condition called aortic aneurysm — a swelling in the aorta that can lead to tear or rupture of this major artery and possibly death. Cardiologists often discover a swollen aorta by chance when testing for something else, when the aorta is already at a life-threatening stage. The team envisioned a machine learning model that can detect enlarged aortas in MRIs at a much earlier stage.
They turned to neural networks, a kind of computer algorithm modeled on how the human brain learns. They first measured the diameters of the ascending and descending aorta by hand in about 100 MRI images, and then used that data to “train” their model to make these measurements automatically, in a process called deep learning. The team applied their model to all of the aortic MRI data in the UK Biobank — nearly 4.5 million images, from more than 43,000 participants.
Next, the researchers analyzed the aortic measurements along with genetic data from the UK Biobank participants, and found more than 100 locations in the genome connected with variation in the aorta. This finding points to genes that scientists could study further to better understand aortic size and risk of aortic aneurysm.
They also developed a polygenic score that distilled down 89 genetic variants associated with enlarged aortas to a single number — a risk score that, together with clinical information, may suggest how likely a person is to develop an aneurysm, even before any symptoms emerge.
Ellinor’s team has used this approach to study a wide range of other cardiac features, such as the structure of the left and right sides of the heart as well as T1 time, a measure of scarring in the heart. They have also found genetic factors linked to aortic distention, a measure of blood flow, and to key features in heart rate monitor readouts. Each time, they found genes for future study and developed polygenic scores to help aid diagnoses.
Ellinor hopes that approaches like these will not only help researchers learn more about the fundamental mechanisms of disease, but also contribute to early disease detection by automating the interpretation of MRI or echocardiogram data, or flagging abnormal images for further analysis.
“We’d like to see this brought back to the clinic, and that requires lots of work,” Ellinor said. “But we’ve built this tool, and it’s showing a lot of promise.”

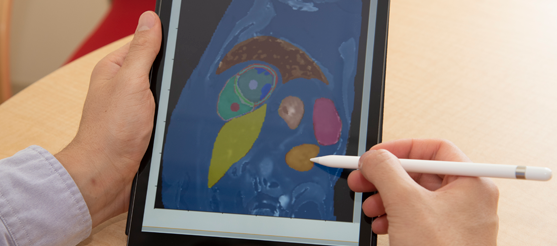
Victor Nauffal, a cardiologist on Ellinor’s team, uses an iPad to annotate structures in an MRI image of the heart by hand.
Credit: Allison Dougherty

Nauffal at work annotating MRI images. These annotations can be used to train machine learning models to recognize different structures of the heart automatically.
Credit: Allison Dougherty
Into the clinic
Doctors assess a patient’s risk for heart disease by looking at certain risk factors, including body mass index (BMI). But Saaket Agrawal wondered whether fat distribution at different locations around the body could be linked to heart disease risk, independent of a patient’s BMI. As a Sarnoff Fellow in Amit Khera’s lab at the Broad, Agrawal — who is also a fourth-year medical student at Northwestern University — partnered with machine learning scientists on the ML4H team, including Marcus Klarqvist, to build algorithms that could analyze whole-body MRI images from thousands of individuals to quantify body fat distribution.
 Saaket Agrawal
Saaket AgrawalThe team built a deep learning model to analyze MRI images and determine fat volumes in three areas of the body — the hips and thighs, in the belly near the skin, and deeper in the belly. Using this tool, they looked for links between body fat distribution and heart disease risk.
“Machine learning enabled us to accurately estimate fat volumes from tens of thousands of MRIs,” Agrawal said. “Increasing sample size in this way helped us understand associations with heart disease and carefully study the genetic underpinnings of body fat distribution.”
Agrawal’s team found that — for individuals of a given BMI — fat in each area of the body had unique associations with risk of coronary artery disease. Fat deeper in the belly was associated with increased risk of coronary artery disease. Patients who had fat in their hips and thighs, on the other hand, had reduced risk of the disease. The researchers also showed that different genes are associated with these three patterns of fat distribution, and that polygenic risk scores for each region of body fat may help indicate a person’s risk of coronary artery disease.
To broaden their method beyond MRI images, Agrawal’s team wondered if they could determine specific fat volumes from just the outline, or silhouette, of an individual. The team built another deep learning model using the silhouette of each individual instead of their full MRI image, and found that it predicted specific areas of fat accurately.
In the future, Agrawal says doctors might be able to estimate these measures of fat distribution from a person’s smartphone-captured silhouette and combine this information with existing clinical knowledge to better predict a person’s risk of cardiovascular disease. They could also monitor the patient to see how their fat distribution — and disease risk — change over time, and recommend appropriate medications or lifestyle modifications.
“Ultimately, this could be another tool in a cardiologist’s toolbox to help them customize medical interventions and lifestyle counseling to the individual in a way that’s most useful for that patient,” Agrawal said.
Cautious optimism
Others at the Broad are using machine learning to take advantage of a major source of biological and clinical data: electronic health records. The work could potentially allow doctors to predict a patient’s future risk of disease from their current health status and medical history, and determine the best treatment accordingly.
 Sarah Urbut
Sarah Urbut“In a 15 or 20 minute visit with a patient, you can’t always access the data from all the interactions they’ve had with the healthcare system,” said Sarah Urbut, a postdoctoral researcher in the laboratory of Pradeep Natarajan at the Broad and a cardiology fellow at Mass General who is developing statistical methods to predict the effect of multiple genes on lipid levels. “It would be awesome if I could incorporate not just their blood pressure that was measured on a certain visit, but also aggregated blood pressure readings over time, in combination with genetic information, into a risk predictor that could tell me which clinical trials might help my patient.”
Broad scientists are now laying the groundwork for this practice, by first filling in gaps in electronic health records. Lab results, imaging data, and doctors’ notes in a patient’s health record can be misclassified, incomplete, or biased because, for instance, they are often generated when the patient is unwell.
To recover missing data in electronic health records, a team led by Batra, together with Mass General cardiologist Steven Lubitz and Broad postdoctoral researcher Shaan Khurshid and Broad machine learning engineers Chris Reeder and Pulkit Singh, are harnessing a machine learning approach called natural language processing. These technologies use statistical methods and deep learning models to interpret text — such as doctors’ notes — using rules, and make inferences about meaning or intent, similar to the way humans do. In April 2022, they also built a pipeline for processing health record data that could help reduce bias in electronic health records by extracting more kinds of data from patient visits. Their pipeline, called JEDI, could help researchers develop more accurate and generalizable machine learning models using electronic health records.
Batra says there’s a long way to go before these methods reach patients. He hopes their work will inspire others to see the potential in passively accumulated data such as medical records, and notes the responsibility researchers have to share their work equitably and broadly.
“It’s critical to our mission that this work ultimately benefits patients,” he said.
The work ahead
Ellinor hopes to see clinicians one day adopting machine learning algorithms in their practices, but he cautions that these methods are still in their infancy, and that scientists will need to thoroughly test them before they are adopted widely. Furthermore, much of the data used to build these algorithms comes from biobanks, which offer scientists thousands or millions of data points but are not representative of every population.
“One of the biggest challenges of applying these tools to the clinic is not making the machine algorithms, but rigorously and repeatedly testing the data and tools to minimize errors and bias,” he said. “I think we have to be very cautious right now.”
Philippakis said doctors don’t currently use many software tools to support their decision making, and may not yet be comfortable with an algorithm recommending a diagnosis. He hopes to see the development and implementation of regulatory frameworks that might help facilitate the adoption of these methods by clinicians.
Even still, Philippakis and Ellinor consider it a success that cardiologists and computer scientists have figured out how to work together, despite some key differences between the two communities. Philippakis says that software developers are used to rapid iterative cycles, while medicine has less room for trial and error. “In the beginning, I wouldn’t have been at all surprised if it just fell apart and nothing got done,” he said.
But he adds that though building these communities takes time, he has seen how these groups are united by the promise of improving health and medicine through data. “Creating a new generation of tools that help doctors make better decisions through data is not a small opportunity,” Philippakis said. “In some ways, I think this is the journey of our time in medicine.”
MRI image in header reproduced by kind permission of UK Biobank©. This research has been conducted using the UK Biobank Resource under Application Number 7089.
A community of computer scientists and physician-scientists at Broad is applying computational approaches to patient data to gain new insights into heart disease.
